As an A-Level student, this placement was an eye opening experience into the world of science.
I discovered how physics can be applied to study lipids and how different scientific research is to what I expected. I was excited to find out that there is a significant amount of computational research in the Department of Physics at King’s College London.
My whole project was computational. During my project, I used molecular dynamics simulations to investigate how lipids interact with each other and if they would aggregate to form a micelle. Molecular dynamics simulations deal with classical mechanics and Newton’s 2nd law.
I studied a phosphocholine lipid, which is similar to that found in the cell membrane, that self-assembles due to its hydrophobic and hydrophilic structure.
My project included the following steps:
- Set up the simulation containing 19 lipids and 35,150 water molecules, resulting in a 28mM concentration solution of lipids. The aqueous phased consisted of 150mM salt concentration with 96 Na+ ions and 96 Cl-ions
- Minimise the molecules (lowest energy conformation)
- Equilibrate the simulation
- Run molecular dynamics simulation for about 100 ns.
- Analyse simulation using Python; checking the size and shape and aggregation number of time.
My simulation gave no obvious trend with number of aggregates over time. This was rendered at the start and end of the simulation (see figures below), showing us no micelle forming, but clusters aggregating and dissociating throughout. This means no micelle had formed due to one of two reasons: (i) simulation did not run long enough to form a micelle; or (ii) 19 lipids are not enough to form a stable micelle.
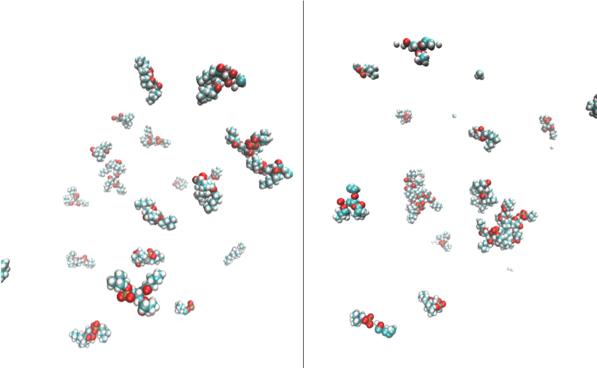
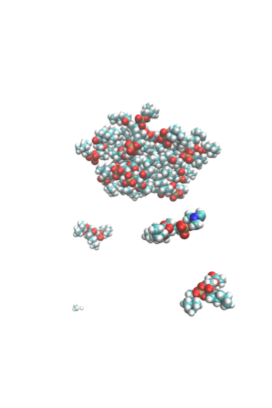
In order to investigate if a micelle of 19 lipids was stable at this concentration, Demi Pink, the PhD student who I worked with during my placement, simulated a pre-assembled micelle of 19 lipids to see if it would dissociate. This simulation showed that overall the micelle (see snapshot to the right) does not dissociate, but a few lipids do disassociate from the aggregate and become monomers in solution. So I believe that my simulation would’ve formed a micelle if the simulation had been extended.
I have achieved this all in 2 weeks and it was all thanks to Dr. Chris Lorenz and Demi for volunteering to do this with me and giving me their precious time, which resulted in an amazing opportunity for me.
This placement gave me an insight into science which is perfect to hold onto with my upcoming university application.
I also thank the Department of Physics at King’s College London for being so welcoming.
— Mariam Yassine


Congratulations on your well written summary of your experience. Best wishes as you continue your pursuit of knowledge!
احسنتي الله يوفقك لما فيه صباحك وصلاح البشرية مزيد من التقدم الحضري بالتوفيق من الله