Machine learning has the potential to revolutionize the design of new functional materials, but building the large and diverse databases of molecules required for training these methods remains a challenge. Automated computational chemistry workflows have become an essential tool in this data-driven search for new materials with novel properties, providing a means of creating and curating molecular databases without significant user input. This helps address concerns about data provenance, reproducibility, and replicability.
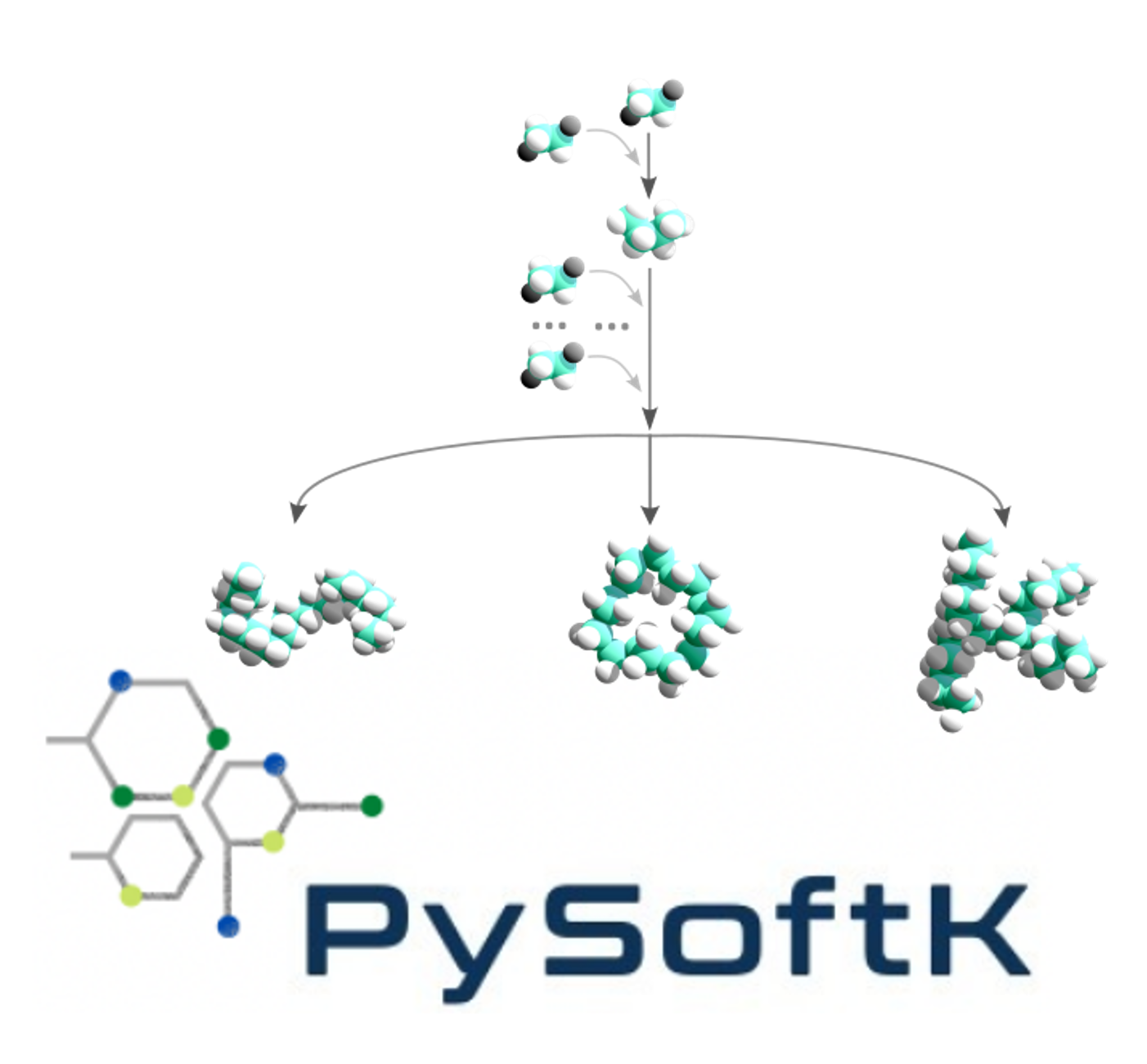
Our group (with a load of help from Alejandro Santana Bonilla) has developed PySoftK, a versatile and flexible software package that provides automated computational workflows for creating, modeling, and curating libraries of polymers with minimal user intervention. PySoftK is available as an efficient and easily installed Python package, and its key features include the ability to generate a wide range of polymer topologies and fully parallelized library generation tools.
With its ability to generate, model, and curate large polymer libraries, PySoftK is expected to support the discovery of functional materials in nano- and biotechnology. Automated computational chemistry workflows like PySoftK are poised to play a crucial role in the data-driven design of new functional materials and the creation of the molecular databases required for machine learning applications.